Q&A: Research at Washington University in St. Louis is Aimed at Developing a Precision Medicine Approach to Fight Neurological Diseases
Blog: The Tibco Blog
Reading Time: 3 minutes
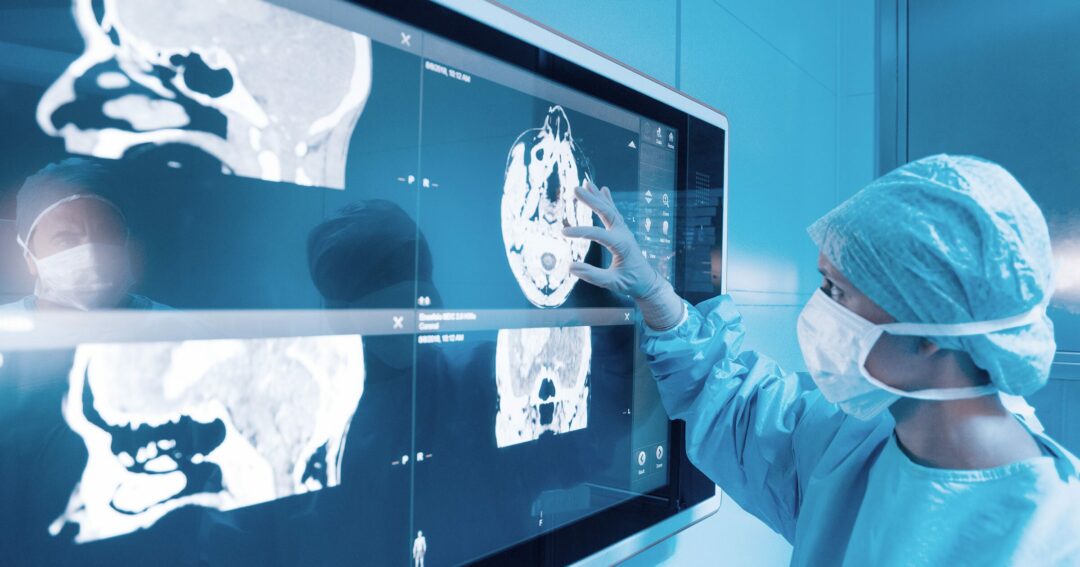
Researchers at Washington University School of Medicine in St. Louis are developing a precision medicine approach to fighting neurological diseases, screening thousands of cell mutations to find and alter the ones that cause illness. Washington University in St. Louis presented an overview of the work at TIBCO NOW 2020. We sat down with William Buchser, assistant professor of genetics and director of the F.I.V.E. @ MGI facility, and Jack Bramley, a scientist in the Buchser Lab, to get a closer look.
TIBCO: What does your research entail?
William Buchser: There are a number of different therapies out there used to treat genetic diseases. One of the newest is gene therapy, utilizing a virus to “fix” a gene because it was broken. As that tool becomes more available therapeutically, then the question becomes, “What gene do we fix?” That’s where we come in—to figure out what gene is broken so that we can pass that information on to scientists to conduct additional research that may lead to the development of gene therapy to fix the problem. Finding these mutations is called functional genomics, and we employ CRISPR to generate mutations in human cell lines that we screen for microscopic markers of disease.
TIBCO: What are some of your challenges with regard to the data in your research?
WB: We humans have about 20,000 genes in our genome, and the DNA sequences of those genes can vary from person to person. If you consider small changes in the letters of those genes (called variants), then each of us has about a million of these compared with any other person. Someone with a disease has almost a million variants that are not harmful and only a handful that are. The challenge is having a tool to manage the datasets and large numbers of combinations of variants that we are testing and keep track of how the data moved through the analysis system in a natural way.
TIBCO: In addition to variants, what other data points do you look at?
JB: We have a number of different data sources that we use. We use confocal imaging to take pictures of millions of cells and figure out—through predictive analytics—if that cell is harboring a disease-causing variant. The dataset considers these cells as a matrix of features. We then get sequencing data for particular cells chosen for further analysis, and there is experimental metadata connected with the cells as well.
TIBCO: How do you use TIBCO software?
JB: We use TIBCO Spotfire to bring in our cell-based datasets and run it through quality control procedures. We also use it to generate our early machine learning models quickly, so we can start selecting cells for picking as soon as possible. We also use TIBCO Data Science Workbench (Statistica) to generate more advanced models to predict which cells are perturbed by the mutations. Once sequence data is available, Spotfire Data Canvas is used to bring all the data together and make our final analyses.
TIBCO: How many people are using the tools?
WB: Seven full-time members of the lab and five undergraduate students who rotate through the lab. We show the students how to use the tool, and then they can sit down in front of a dataset and actually see the data, manipulate it, and see the results. It is wonderful for WashU and our group to have access to these tools and technologies that assist with cutting-edge research but also expose students to the tools and facilitate their exploration of the technology.
Researchers at Washington University School of Medicine in St. Louis are developing a precision medicine approach to fighting neurological diseases, screening thousands of cell mutations to find and alter the ones that cause illness.
Click To Tweet
To learn more about how universities and other academic institutions are leveraging TIBCO products, check out the TIBCO Academic Alliance.
Leave a Comment
You must be logged in to post a comment.